Full text loading...
Plant disease detection represents a tremendous challenge for research and practical applications. Visual assessment by human raters is time-consuming, expensive, and error prone. Disease rating and plant protection need new and innovative techniques to address forthcoming challenges and trends in agricultural production that require more precision than ever before. Within this context, hyperspectral sensors and imaging techniques—intrinsically tied to efficient data analysis approaches—have shown an enormous potential to provide new insights into plant-pathogen interactions and for the detection of plant diseases. This article provides an overview of hyperspectral sensors and imaging technologies for assessing compatible and incompatible plant-pathogen interactions. Within the progress of digital technologies, the vision, which is increasingly discussed in the society and industry, includes smart and intuitive solutions for assessing plant features in plant phenotyping or for making decisions on plant protection measures in the context of precision agriculture.

Article metrics loading...

Full text loading...
Literature Cited


Data & Media loading...
Supplemental Material
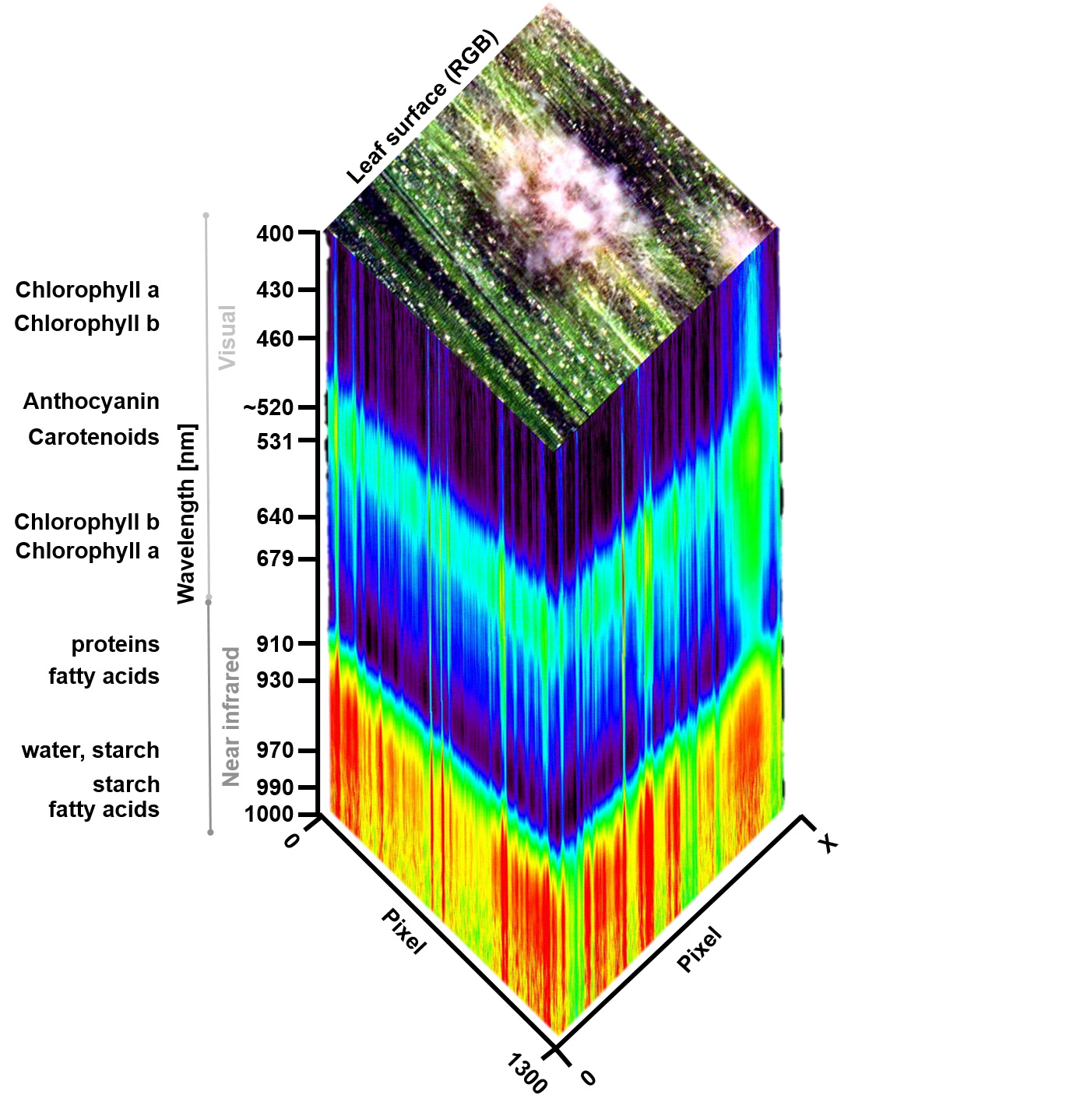
Supplemental Figure 1. Hyperspectral imaging data cube of a barley leaf, diseased with powdery mildew, caused by Blumeria gramins f.sp. hordei. A hyperspectra ldata cube contains the spatial information in two dimensions and can be visualized as a pseudo-RGB image. Spectral reflectance properties for each image pixel are available from 400 to 1,000 nm and are shown as an intensity image (rainbow colors). Wavelength with maximal absorbance in the visible range and near-infrared range of different pigments and other leaf chemical compounds are indicated. Download Supplemental Figure 1 as a PDF.