Full text loading...
Trichoderma species are widely used in agriculture and industry as biopesticides and sources of enzymes, respectively. These fungi reproduce asexually by production of conidia and chlamydospores and in wild habitats by ascospores. Trichoderma species are efficient mycoparasites and prolific producers of secondary metabolites, some of which have clinical importance. However, the ecological or biological significance of this metabolite diversity is sorely lagging behind the chemical significance. Many strains produce elicitors and induce resistance in plants through colonization of roots. Seven species have now been sequenced. Comparison of a primarily saprophytic species with two mycoparasitic species has provided striking contrasts and has established that mycoparasitism is an ancestral trait of this genus. Among the interesting outcomes of genome comparison is the discovery of a vast repertoire of secondary metabolism pathways and of numerous small cysteine-rich secreted proteins. Genomics has also facilitated investigation of sexual crossing in Trichoderma reesei, suggesting the possibility of strain improvement through hybridization.

Article metrics loading...

Full text loading...
Literature Cited


Data & Media loading...
Supplemental Material
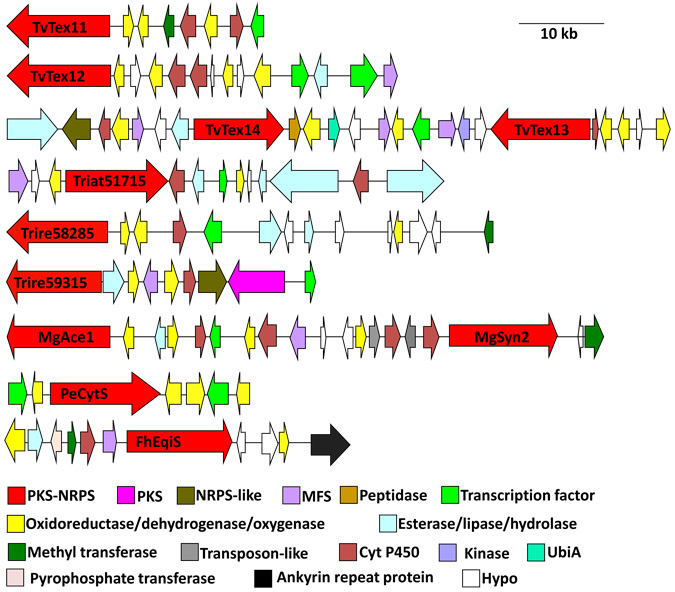
Download all Supplemental Materials as a PDF. Includes Supplemental Table 1: A summary of some features of the sequenced genomes and Supplemental Figure 1: PKS-NRPS gene clusters of Trichoderma spp. (also reproduced below). Supplemental Figure 1: PKS-NRPS gene clusters of Trichoderma spp.