Full text loading...
Ruminant production systems face significant challenges currently, driven by heightened awareness of their negative environmental impact and the rapidly rising global population. Recent findings have underscored how the composition and function of the rumen microbiome are associated with economically valuable traits, including feed efficiency and methane emission. Although omics-based technological advances in the last decade have revolutionized our understanding of host-associated microbial communities, there remains incongruence over the correct approach for analysis of large omic data sets. A global approach that examines host/microbiome interactions in both the rumen and the lower digestive tract is required to harness the full potential of the gastrointestinal microbiome for sustainable ruminant production. This review highlights how the ruminant animal production community may identify and exploit the causal relationships between the gut microbiome and host traits of interest for a practical application of omic data to animal health and production.

Article metrics loading...

Full text loading...
Literature Cited


Data & Media loading...
Supplemental Material
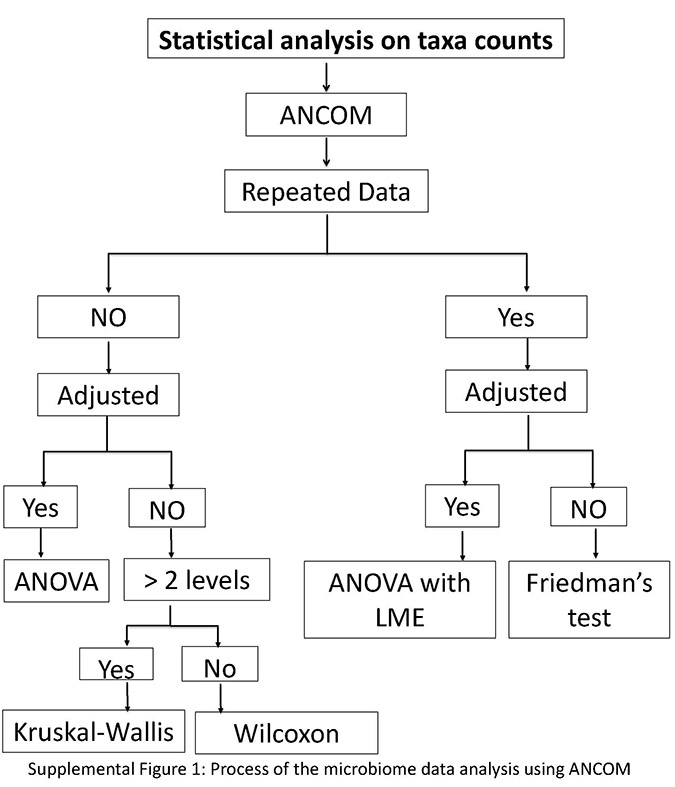
Download Supplemental Figures 1-2 as a PDF, or see below. Supplemental Figure 1. Supplemental Figure 2.